|
Introduction
Research in the Robinson
group centers on three areas in biochemical engineering:
understanding and controlling protein aggregation; cellular
mechanisms controlling protein quality and human disease; and
overcoming obstacles to expression and characterization of
G-protein coupled receptors. In each of these areas, we use
the tools of molecular and cellular biology, biochemistry, and
biophysics, combined with systems biology, mathematical
modeling, and engineering analysis, to develop an improved
understanding of biological systems. Using this knowledge, we
carry out molecular and cellular engineering to develop
improved methods, products, and tools for biotechnology,
medical, and research applications.

Identifying and Overcoming
Obstacles to GPCR Expression and Characterization
Human
G-protein coupled receptors (GPCRs) represent the largest
family of integral membrane receptors that are involved in
intercellular communication in response to diverse external
stimuli. Upon ligand
binding to GPCRs, a signal is initiated intracellularly via
interaction with the GTP-binding protein (G protein) to
further transmit cellular responses. GPCRs play a myriad
number of physiological roles in humans, leading to a
substantial pharmacological interest in modulating this
signaling activity via novel pharmaceutically active
molecules. Indeed, 30-50% of currently marketed drugs are
thought to be act through the direct and indirect modulation
of GPCR activity.
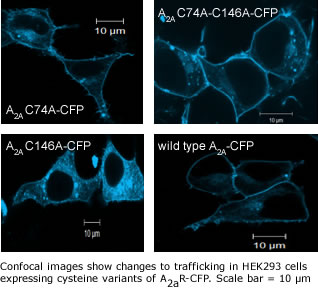
Although cloning of a mammalian GPCR (β2 adrenergic
receptor) was first achieved in the mid-1980s, the first
non-rhodopsin GPCR structure only became available almost 20
years later (2007). There are approximately 360 non-sensory
human GPCRs (of ~800-900 total predicted GPCRs), with ligands
identified or predicted for only ~2/3 of the non-sensory
proteins. Many acute and chronic disease states are linked to
GPCR function or malfunction, and 30-60% of commercially
available drugs interact with a GPCR, making them targets for
nearly 40% of drug discovery efforts worldwide, yet these
drugs target less than 10% of all GPCRs. However, efforts to
better understand GPCR ligand specificity, structure,
stability, and assembly are hampered by the difficulties
associated with producing these integral membrane proteins.
Our research in this area includes re-engineering of
the recognition sequence of GPCRs to serve as cellular
sensors, improving expression of GPCRs for drug screening and
crystallization, and biophysical characterization of
ligand-receptor and membrane-receptor interactions.
Recent
and Representative Publications:
- Naranjo,
AN, A Chevalier, GD Cousins, E Ayettey, EC McCusker, C
Wenk, AS Robinson (2015) Conserved disulfide bond is not
essential for the adenosine A2A receptor: extracellular
cysteines influence receptor distribution within the cell
and ligand-binding recognition, BBA
Biomembranes, 1848: 603-614 Available on-line Dec 5
2014 DOI: 10.1016/j.bbamem.2014.11.010
- Blocker,
KM, ZT Britton, AN Naranjo, PM McNeely, CL Young, AS
Robinson (2015) Recombinant G protein-coupled receptor
expression in Saccharomyces
cerevisiae for protein characterization, in
“Membrane Proteins – Production and Function
Characterization”, Methods Enzymol., 556:165-83. doi: 10.1016/bs.mie.2014.12.025
- McNeely,
P.M., Naranjo, A.N., and A.S. Robinson (2012)
Structure-function studies with G-protein coupled
receptors as a paradigm for improving drug discovery and
therapeutic development, Biotechnology
Journal, 7(12): 1451-1461. DOI:
0.1002/biot.201200076
- O’Malley,
Michelle A., Matthew E. Helgeson, Norman J. Wagner, Anne
S. Robinson (2011) “Toward Rational Design of Protein
Detergent Complexes: Determinants of Mixed Micelles that
are Critical for the in
vitro Stabilization of a G-protein Coupled
Receptor”, Biophysical Journal, 101 (8): 1938-1948. DOI:
10.1016/j.bpj.2011.09.018 PMID: 22004748
Collaborators:
Bramie
Lenhoff
Department of Chemical Engineering
University of Delaware
http://www.che.udel.edu/directory/facultyprofile.html?id=252
Wilfred
Chen
Department
of
Chemical Engineering
University of Delaware
http://www.che.udel.edu/directory/facultyprofile.html?id=25009
Funding:
National
Science
Foundation 1263768 (PI: W. Chen)
Collaborative
Proposal: Exploiting synthetic GPCRs and mating factors as
extracellular sensors for substrate-dependent assembly of
complex cellulosomes

Understanding
and Controlling Protein Aggregation
Aggregation
is a long-standing in
vitro and in
vivo obstacle
for studying proteins; it serves as an irreversible,
off-pathway process during protein folding, and it is a
ubiquitous problem throughout commercial manufacture of
protein-based biotechnology products. This is a potentially
debilitating setback from a structural biology and protein
design perspective, as it can limit the ability of
scientists to produce sufficiently high quantities of
purified material that are required for subsequent sample
preparation and for biophysical characterization. It also
greatly limits biotechnology product discovery and
development. From a discovery perspective, only those
candidate molecules that can be readily expressed and
(re)folded to active forms can be included in screens for
improved or novel structure and function.
As
these stresses cannot be avoided altogether practically,
an attractive alternative strategy is to engineer protein
sequence/structure to be less susceptible to aggregation.
If this can be done within a mechanistic context of
protein aggregation, then it may be more easily
generalized beyond model systems. Therefore, an overall
motivation for the collaborative research with the Roberts
lab is to provide mechanistic yet practical computational
tools and design paradigms for rational protein design to
impart resistance to aggregation, and in the longer term
to combine them with algorithms focused on maintaining or
imbuing new protein function.
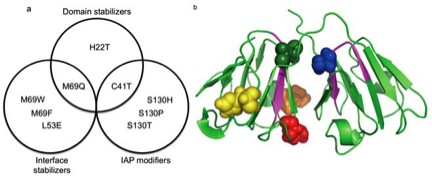
(a)
Summary of designed point-mutants for gDCrys based on
category of design strategy. (b)Three dimensional structure
of gDCrys (PDB file: 1HK0) illustrating each variant site;
H22 (yellow), C41 (green), L53 (red), M69 (orange), and S130
(blue). From Sahin, E et al. (2011) Computational Design and
Biophysical Characterization of Aggregation-Resistant Point
Mutations for gD Crystallin Illustrate a Balance of
Conformational Stability and Intrinsic Aggregation
Propensity, Biochemistry
50, 628-639.
Recent and Representative
Publications:
- Wu,
H, K Truncali, J Ritchie, R Kroe-Barrett, S Singh, AS
Robinson, and CJ Roberts (2015) Weak protein
interactions and pH- and temperature-dependent aggregation
of human Fc1, mAbs,
in press
- Wu,
H., Rachel Kroe-Barrett, Sanjaya Singh, Anne S. Robinson,
Christopher J. Roberts (2014) Competing aggregation
pathways for monoclonal antibodies, FEBS
Letters 588(6): 936-941. http://dx.doi.org/10.1016/j.febslet.2014.01.051
- JA
Costanzo, CJ O’Brien, K Tiller†, E Tamargo†,
AS Robinson, CJ Roberts, and EJ Fernandez (2013)
Computational Design to Control Protein Aggregation Rates
Through Conformational Stability, Protein
Eng, Des, & Sel, 27 (5): 157-167
10.1093/protein/gzu008
Collaborator:
Chris
Roberts
Department of Chemical Engineering
University of Delaware
http://www.che.udel.edu/directory/facultyprofile.html?id=12564
Funding:
NSF
1264554 (PI: Roberts)
GOALI: Collaborative Proposal: Mechanistic Design of
Aggregation Resistance in Multi-Domain Proteins

Cellular Mechanisms
Controlling Protein Quality and Human Disease
Cells
are inherently robust to stochastic perturbations, and have
evolved to recover readily from short-term exposure to heat,
pH changes, and nutrient deprivation during times of stress.
This process, termed the stress response, is important in a
wide range of basic research and commercial applications
since cell growth rates, production of metabolites, and
protein expression are all affected by the stress response.
Moreover, accumulation of toxic metabolic products or
unfolded proteins can in turn induce the stress response,
implicated in a number of human diseases. Our two areas of
research are:
- Understanding
the effects of these cellular control mechanisms of protein
expression levels and protein quality (activity,
post-translational modifications).
- Determining
how loss of cellular control may lead to a disease state
During
protein expression, the stress response to unfolded protein
accumulation, termed the unfolded protein response (UPR),
resulting in low protein yields during heterologous protein
expression. A systematic approach to improving protein
production involves understanding on a molecular level how
cell regulation mechanisms respond to heterologous protein
expression. Key issues include how to maintain reasonable
cell health, yet obtain high protein yields and what
interactions promote or stabilize formation of active
protein with correct post-translation modifications.
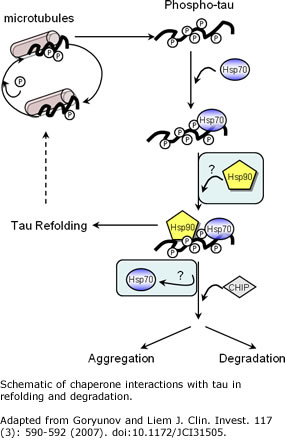
Hallmarks
of the disease state in the Alzheimer brain, and of
several other neurodegenerative diseases including
corticobasal degeneration, frontotemporal dementia, and
parkinsonism linked to chromosome 17, are the
hyperphosphorylation of tau and subsequent formation of
insoluble tau aggregates (neurofibrillary tangles or
paired helical filaments). In healthy cells, the tau
protein binds to and stabilizes microtubules, and is
abundant. It is not yet clear whether the problem in these
diseases is a loss of tau function (e.g. loss of
microtubule stability), or an inherent toxicity of the tau
tangles. Major research questions include determining the
biochemical and cellular pathways that drive tau
homeostasis, including degradation and refolding pathways,
and which steps in the tau pathways are the best targets
for therapeutic intervention.
Recent and Representative
Publications:
- Young,
CL and AS Robinson* (2014) Protein Folding and
Secretion: Mechanistic Insights Advancing Recombinant
Protein Production in S.
cerevisiae, Current Opinion in Biotechnology 30:
168-177. DOI: 10.1016/j.copbio.2014.06.018
- Spatara, ML and Robinson,
AS* (2010) “Transgenic mouse and cell culture models
demonstrate a lack of mechanistic connection between
endoplasmic reticulum stress and tau dysfunction” Journal
of Neuroscience Research, 88(9):1951-61. PMID:
20143409

|